CausalCCC
FROM CELL-CELL COMMUNICATION METHODS TO INTEGRATIVE CAUSAL PATHWAYS
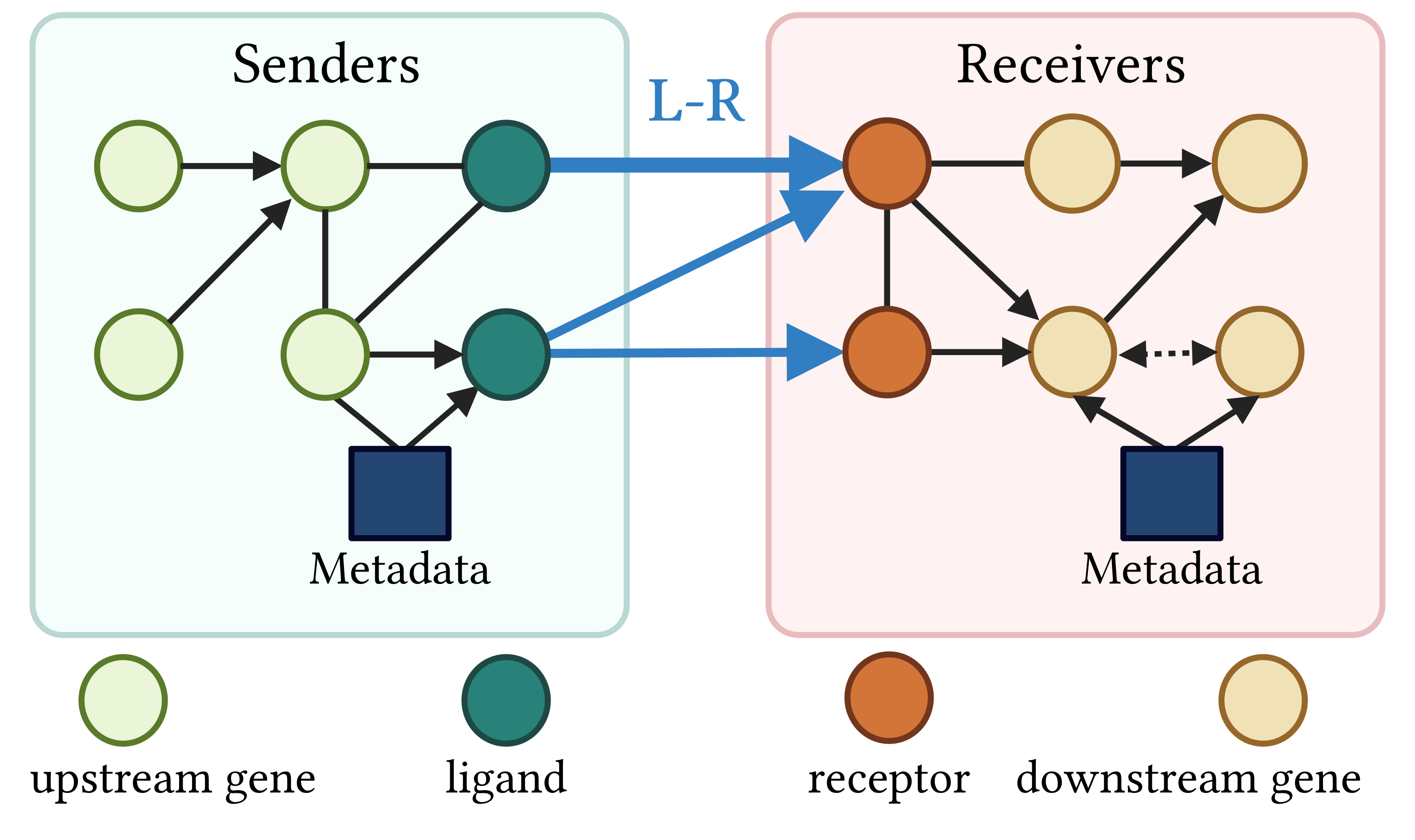
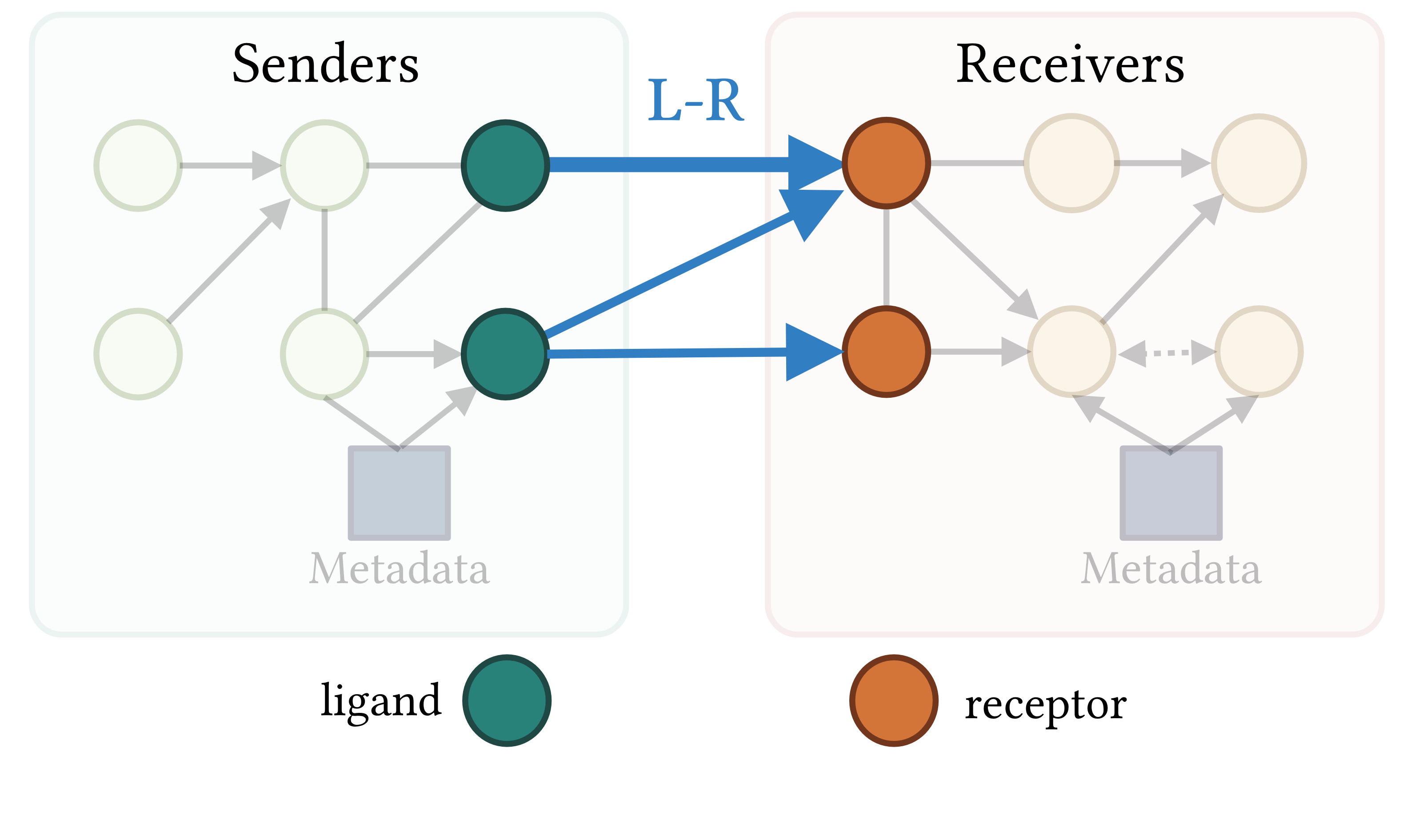
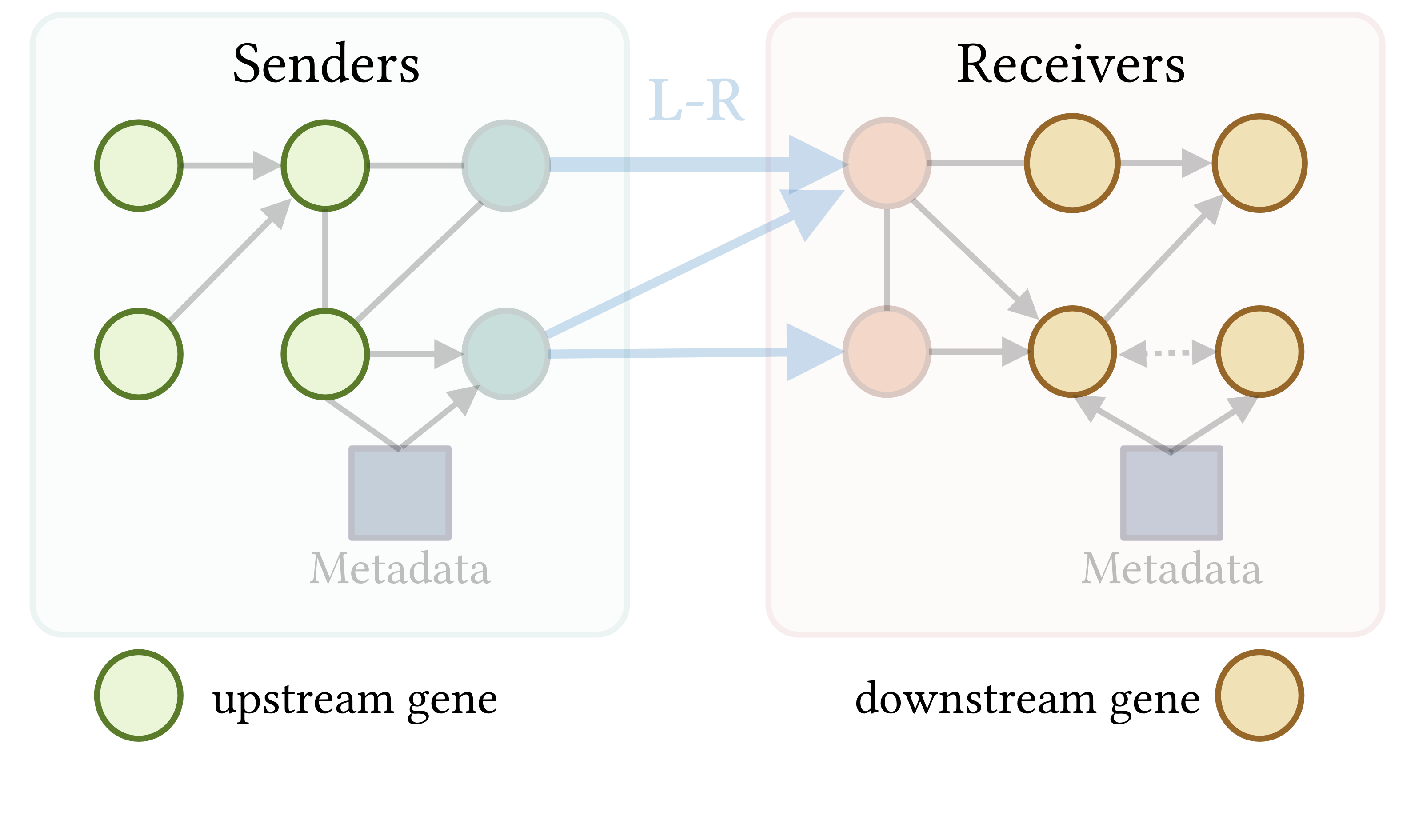
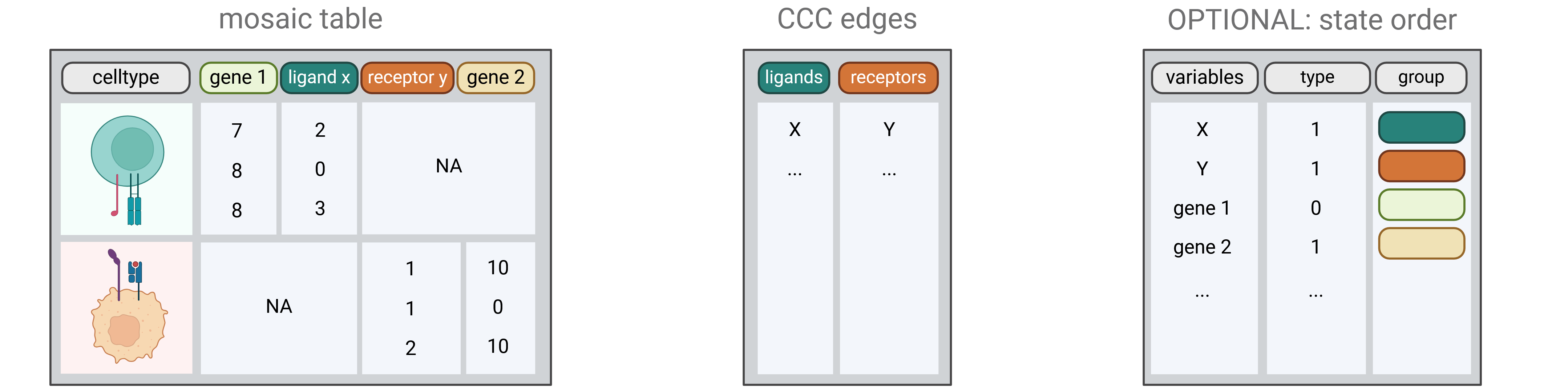
CausalCCC offers an interactive webserver solution to reconstruct gene interaction pathways across interacting cell types, downstream of the usual processing of single-cell RNAseq objects. This allows you to keep total control of your single-cell object.
See tutorials.
See publication here :
paper
See detailed CausalCCC best practice here :
tutorial